See what you are missing - try a Literature Lab™ PLUS analysis on your next dataset
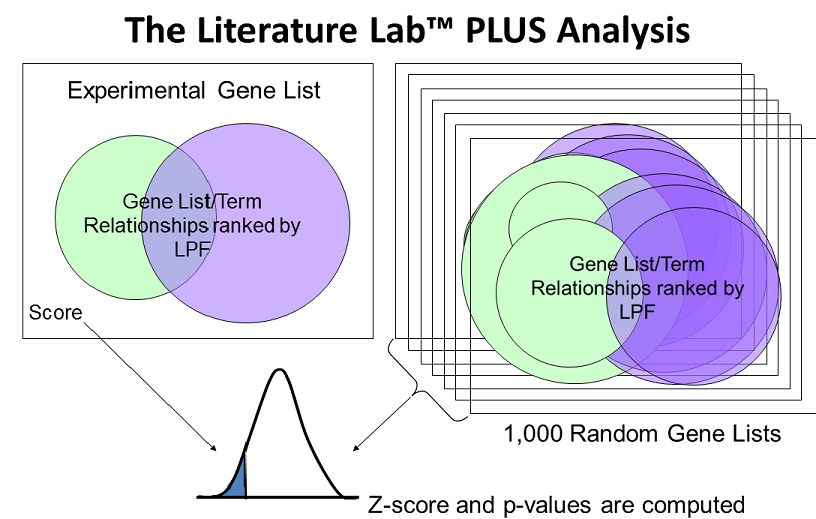
Literature Lab™ PLUS analyzes the relationships of your datasets to biological concepts in the literature, using a 1,000 RGS statistical analysis described below.
Unlike virtually all other enrichment tools, Literature Lab™ PLUS does not depend on fixed databases of gene lists or a priori determinations. No matter how good these enrichment tools are, and no matter how fancy their algorithms, they still base their analysis on…other gene lists!
Literature Lab™ PLUS respects the uniqueness of each gene list you submit for analysis and returns a consistent and unique result accordingly. By looking in real time at the relationship of each gene list to biological concepts in the literature, Literature Lab™ PLUS identifies critical relationships that the other approaches miss.
Literature Lab™ PLUS is the most powerful tool available for transcriptomic functional analysis. It identifies significant associations in 25 domains of biology and biochemistry, including pathways, diseases, cell biology, chemicals/drugs, cancer drugs, inhibitors, metabolites and physiology.
At the core of the Literature Lab™ database is the Acumenta Biotech Gene Thesaurus™, a repository of gene, protein and pathway nomenclature gathered from the major genomic databases, and human-curated to produce searches combining high precision and high recall. The Gene Thesaurus™ is unique because it is constantly updated on the Literature™ Lab platform, and human curation resolves ambiguous terminology, alias redundancy and generic terms.
the Literature Lab™ PLUS Analysis.
The LPFs (log of the product of frequency) of the genes in the input set are computed with each of the 86,000 terms in the Literature Lab™ database. This analysis is also generated on each of 1,000 random gene sets (RGS). The LPFs for the input data set are compared to the 1,000 RGS and statistically significant associations unique to each gene list analyzed, are identified.
A Literature Lab™ PLUS analysis on 100 genes is equivalent to 2.4 billion manual searches.
ANALYSIS RESULTS ARE PRESENTED IN A POWERFUL VIEWER.
The Literature Lab™ PLUS Viewer is a point-and-click application that quickly reveals the biological meaning of your gene set. Significantly associated pathways, diseases, chemicals, drugs, cell biology and other biological and biochemical dimensions are shown, accompanied by the genes that drive each association, ranked by their contribution to the association. The Literature Lab™ analysis is 100% transparent -- one click takes you to the abstracts that drive each association. There is no more wondering why, how or from where your gene set enrichment results are produced. The Literature Lab™ database is constantly updated, assuring you that the enrichment of your gene set is based on the latest data.
Compare multiple gene sets.
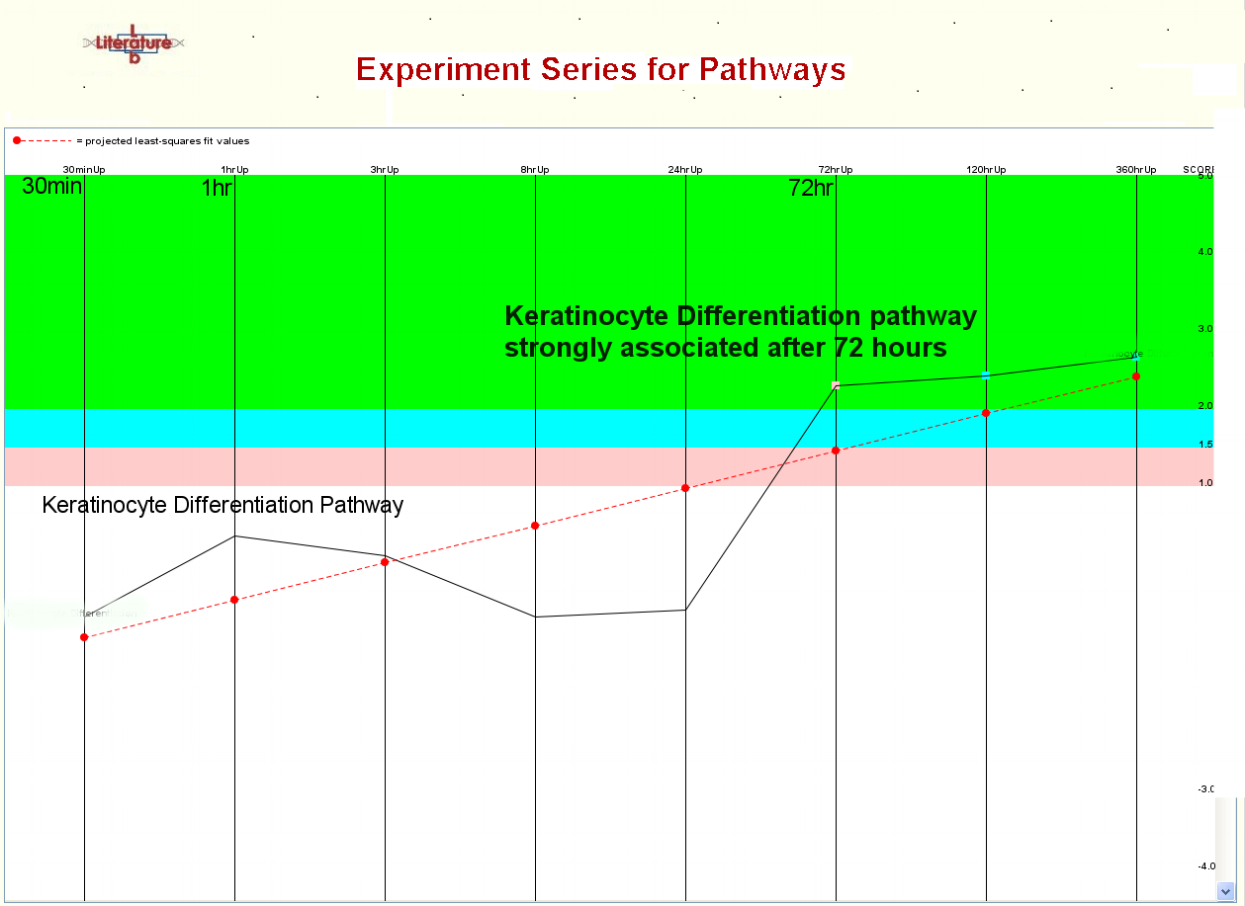
The Literature Lab™ PLUS Viewer compares the results of multiple gene list analyses. What are the biological similarities and differences between two gene sets? The answer is a few clicks away. Time or drug course data? Data from multiple tissue types?...The PLUS Viewer shows the trends, changes, peaks, etc. across multiple gene sets.
ASSOCIATIONS OBTAINED WITH A SINGLE CLICK
IL10 is one of the drivers of this gene list's association with the Acute Inflammatory Response Pathway. This image shows the results of a single-click request for genes associated in the literature with IL10 AND the Acute Inflammatory Response Pathway. Genes in the gene list are displayed in bold blue. Note that the LPF in this example is computed on the selected gene's association with the genes in the literature.
Functional insights, targets revealed.
Quickly make sense out of larger heterogeneous gene lists - Literature Lab™ PLUS runs a hierarchical clustering analysis on the genes and the significantly associated terms to illuminate functional relationships among groups of genes in the gene set.
The Literature Lab™ PLUS clusters are interactive -- clicking on a cell instantly takes you to the abstracts for the gene and the term. There is no more wondering why, how or from where your gene set enrichment results are produced.